Altair/Pandas: TypeError: Cannot interpret 'Float64Dtype()' as a data type
I ran into an interesting problem when trying to use Altair to visualise a Pandas DataFrame containing vaccination rates of different parts of England. In this blog post we’ll look at how to work around this issue.
First, let’s install Pandas, numpy, and altair:
pip install pandas altair numpyAnd now we’ll import those modules into a Python script or Jupyter notebook:
import pandas as pd
import altair as alt
import numpy as npNext, we’ll create a DataFrame containing the vaccinations rates of a couple of regions:
vaccination_rates_by_region = pd.DataFrame([
{"Region": "East Midlands", "Overall": 48.877331},
{"Region": "London", "Overall": 32.58}
])
vaccination_rates_by_region = vaccination_rates_by_region.convert_dtypes()| Region | Overall |
|---|---|
East Midlands |
48.877331 |
London |
32.58 |
Let’s now try to create a bar chart based on this data:
(alt.Chart(vaccination_rates_by_region).mark_bar().encode(
x=alt.X('Region'),
y=alt.Y('Overall', axis=alt.Axis(title='Vaccinations')),
tooltip=[alt.Tooltip('Overall', format=",")])
.properties(width=600))If we run this code, we’ll see the following error:
TypeError Traceback (most recent call last)
~/.local/share/virtualenvs/covid-vaccines-xEbcGJTy/lib/python3.8/site-packages/altair/vegalite/v4/api.py in to_dict(self, *args, **kwargs)
361 copy = self.copy(deep=False)
362 original_data = getattr(copy, "data", Undefined)
--> 363 copy.data = _prepare_data(original_data, context)
364
365 if original_data is not Undefined:
~/.local/share/virtualenvs/covid-vaccines-xEbcGJTy/lib/python3.8/site-packages/altair/vegalite/v4/api.py in _prepare_data(data, context)
82 # convert dataframes or objects with __geo_interface__ to dict
83 if isinstance(data, pd.DataFrame) or hasattr(data, "__geo_interface__"):
---> 84 data = _pipe(data, data_transformers.get())
85
86 # convert string input to a URLData
~/.local/share/virtualenvs/covid-vaccines-xEbcGJTy/lib/python3.8/site-packages/toolz/functoolz.py in pipe(data, *funcs)
625 """
626 for func in funcs:
--> 627 data = func(data)
628 return data
629
~/.local/share/virtualenvs/covid-vaccines-xEbcGJTy/lib/python3.8/site-packages/toolz/functoolz.py in __call__(self, *args, **kwargs)
301 def __call__(self, *args, **kwargs):
302 try:
--> 303 return self._partial(*args, **kwargs)
304 except TypeError as exc:
305 if self._should_curry(args, kwargs, exc):
~/.local/share/virtualenvs/covid-vaccines-xEbcGJTy/lib/python3.8/site-packages/altair/vegalite/data.py in default_data_transformer(data, max_rows)
17 @curried.curry
18 def default_data_transformer(data, max_rows=5000):
---> 19 return curried.pipe(data, limit_rows(max_rows=max_rows), to_values)
20
21
~/.local/share/virtualenvs/covid-vaccines-xEbcGJTy/lib/python3.8/site-packages/toolz/functoolz.py in pipe(data, *funcs)
625 """
626 for func in funcs:
--> 627 data = func(data)
628 return data
629
~/.local/share/virtualenvs/covid-vaccines-xEbcGJTy/lib/python3.8/site-packages/toolz/functoolz.py in __call__(self, *args, **kwargs)
301 def __call__(self, *args, **kwargs):
302 try:
--> 303 return self._partial(*args, **kwargs)
304 except TypeError as exc:
305 if self._should_curry(args, kwargs, exc):
~/.local/share/virtualenvs/covid-vaccines-xEbcGJTy/lib/python3.8/site-packages/altair/utils/data.py in to_values(data)
147 return {"values": data}
148 elif isinstance(data, pd.DataFrame):
--> 149 data = sanitize_dataframe(data)
150 return {"values": data.to_dict(orient="records")}
151 elif isinstance(data, dict):
~/.local/share/virtualenvs/covid-vaccines-xEbcGJTy/lib/python3.8/site-packages/altair/utils/core.py in sanitize_dataframe(df)
334 col = df[col_name].astype(object)
335 df[col_name] = col.where(col.notnull(), None)
--> 336 elif np.issubdtype(dtype, np.integer):
337 # convert integers to objects; np.int is not JSON serializable
338 df[col_name] = df[col_name].astype(object)
~/.local/share/virtualenvs/covid-vaccines-xEbcGJTy/lib/python3.8/site-packages/numpy/core/numerictypes.py in issubdtype(arg1, arg2)
417 """
418 if not issubclass_(arg1, generic):
--> 419 arg1 = dtype(arg1).type
420 if not issubclass_(arg2, generic):
421 arg2 = dtype(arg2).type
TypeError: Cannot interpret 'Float64Dtype()' as a data typeWe can check the types used in our DataFrame by running the following code:
vaccination_rates_by_region.dtypesRegion string
Overall Float64
dtype: objectThe problem is that altair doesn’t yet support the Float64Dtype type.
We can work around this problem by coercing the type of that column to float32:
vaccination_rates_by_region= vaccination_rates_by_region.astype({
column: np.float32
for column in vaccination_rates_by_region.drop(["Region"], axis=1).columns
})And now if we create a chart:
chart = (alt.Chart(vaccination_rates_by_region).mark_bar().encode(
x=alt.X('Region'),
y=alt.Y('Overall', axis=alt.Axis(title='Vaccinations')),
tooltip=[alt.Tooltip('Overall', format=",")])
.properties(width=600))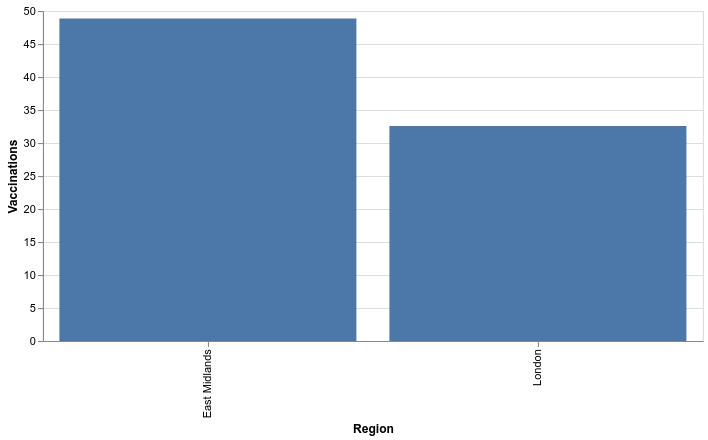
Happy days!
About the author
I'm currently working on short form content at ClickHouse. I publish short 5 minute videos showing how to solve data problems on YouTube @LearnDataWithMark. I previously worked on graph analytics at Neo4j, where I also co-authored the O'Reilly Graph Algorithms Book with Amy Hodler.
